Explanation
Line 0: SELECTED_OUTPUT [number] [description]
SELECTED_OUTPUT is the keyword for the data block. Optionally, SELECTED_OUT, SELECT_OUTPUT, or SELECT_OUT.
number--Positive number to designate this selected-output definition. Default is 1.
description--Optional comment that describes the selected-output data.
-file --Identifier allows definition of the name of the file where the selected results are written. Optionally, file or -f [ ile ].
file name --File name where selected results are written. If the file exists, the contents will be overwritten. File names must conform to operating system conventions. Default is selected.out .
Line 2: -high_precision [( True or False )]
-high_precision --Prints results to the selected-output file with extra numerical precision (12 decimal places, default is 3 or 4). High precision is defined individually for each selected-output file. In addition, the criterion for convergence of the calculations is set to 1 × 10 -12 (default is 1 × 10 -8 ). The convergence criterion also may be set by -convergence_tolerance in KNOBS data block. Default is false at startup. Optionally, high_precision or -h [ igh_precision ].
( True or False )--If true , output is written to the selected-output file with extra decimal places; if false , output to the selected-output file is written with normal precision. If neither true nor false is entered on the line, true is assumed. Optionally, t [ rue ] or f [ alse ].
Line 3: -reset [( True or False )]
-reset --Resets all identifiers listed in Lines 4-18 to true or false . Optionally, reset or -r [ eset ].
( True or False )--If true , identifiers on Lines 4-18 are set to true; if false , identifiers on Lines 4-18 are set to false. If neither true nor false is entered on the line, true is assumed. Optionally, t [ rue ] or f [ alse ].
Line 4: -simulation [( True or False )]
-simulation --Prints simulation number, or for advective-dispersive transport calculations, the sequence number of the advective-dispersive transport simulation. Default is true at startup. Optionally, simulation , sim , or -sim [ ulation ].
( True or False )--If true , simulation number is printed to the selected-output file; if false , simulation number is not printed. If neither true nor false is entered on the line, true is assumed. Optionally, t [ rue ] or f [ alse ].
Line 5: -state [( True or False )]
-state --Prints type of calculation to the selected-output file for each calculation. The following character strings are used to identify each calculation type: initial solution, “i_soln”; initial exchange composition, “i_exch”; initial surface composition, “i_surf”; initial gas-phase composition, “i_gas”; batch reaction (including RUN_CELLS), “react”; inverse, “inverse”; advection, “advect”; and transport, “transp”. Default is true at startup. Optionally, state or -st [ ate ].
( True or False )--If true , state is printed to the selected-output file; if false , state is not printed. If neither true nor false is entered on the line, true is assumed. Optionally, t [ rue ] or f [ alse ].
Line 6: -solution [( True or False )]
-solution --Prints solution number used for the calculation for each calculation. Default is true at startup. Optionally, soln , -solu [ tion ], or -soln . Note the hyphen is required to avoid a conflict with the keyword SOLUTION.
( True or False )--If true , solution number is printed to the selected-output file; if false , solution number is not printed. If neither true nor false is entered on the line, true is assumed. Optionally, t [ rue ] or f [ alse ].
Line 7: -distance [( True or False )]
-distance --Prints to the selected-output file (1) the X-coordinate of the cell for advective-dispersive transport calculations (TRANSPORT), (2) the cell number for advection calculations (ADVECTION), or (3) -99 for other calculations. Default is true at startup. Optionally, distance , dist , or -d [ istance ].
( True or False )--If true , distance is printed to the selected-output file; if false , distance is not printed. If neither true nor false is entered on the line, true is assumed. Optionally, t [ rue ] or f [ alse ].
Line 8: -time [( True or False )]
-time --Prints to the selected-output file (1) the cumulative model time since the beginning of the simulation for batch-reaction calculations with kinetics, (2) the cumulative transport time since the beginning of the run (or since -initial_time identifier was last defined) for advective-dispersive transport calculations and advective-transport calculations for which -time_step is defined, (3) the advection shift number for advective-transport calculations for which -time_step is not defined, or (4) -99 for other calculations. Default is true at startup. Optionally, time or -ti [ me ].
( True or False )--If true , time is printed to the selected-output file; if false , time is not printed. If neither true nor false is entered on the line, true is assumed. Optionally, t [ rue ] or f [ alse ].
Line 9: -step [( True or False )]
-step --Prints to the selected-output file (1) advection shift number for transport calculations, (2) reaction step for batch-reaction calculations, or (3) -99 for other calculations. Default is true at startup. Optionally, step or -ste [ p ].
( True or False )--If true , step is printed to the selected-output file; if false , step is not printed. If neither true nor false is entered on the line, true is assumed. Optionally, t [ rue ] or f [ alse ].
Line 10: -pH [( True or False )]
( True or False )--Prints pH to the selected-output file for each calculation. Default is true at startup. Optionally, pH (as with all identifiers, case insensitive).
( True or False )--If true , pH is printed to the selected-output file; if false , pH is not printed. If neither true nor false is entered on the line, true is assumed. Optionally, t [ rue ] or f [ alse ].
Line 11: -pe [( True or False )]
-pe --Prints pe to the selected-output file for each calculation. Default is true at startup. Optionally, pe .
( True or False )--If true , pe is printed to the selected-output file; if false , pe is not printed. If neither true nor false is entered on the line, true is assumed. Optionally, t [ rue ] or f [ alse ].
Line 12: -reaction [( True or False )]
( True or False )--Prints (1) reaction increment to the selected-output file if REACTION is used in the calculation or (2) -99 for other calculations. Default is false at startup. Optionally, rxn , -rea [ ction ], or -rx [ n ]. Note the hyphen is required to avoid a conflict with the keyword REACTION.
( True or False )--If true , reaction increment is printed to the selected-output file; if false , reaction increment is not printed. If neither true nor false is entered on the line, true is assumed. Optionally, t [ rue ] or f [ alse ].
Line 13: -temperature [( True or False )]
-temperature --Prints temperature (Celsius) to the selected-output file for each calculation. Default is false at startup. Optionally, temp , temperature , or -te [ mperature ].
( True or False )--If true , temperature is printed to the selected-output file; if false , temperature is not printed. If neither true nor false is entered on the line, true is assumed. Optionally, t [ rue ] or f [ alse ].
Line 14: -alkalinity [( True or False )]
-alkalinity --Prints alkalinity (eq/kgw, equivalent per kilogram water) to the selected-output file for each calculation. Default is true at startup. Initial value at start of program is false . Optionally, alkalinity , alk , or -al [ kalinity ].
( True or False )--If true , alkalinity is printed to the selected-output file; if false , alkalinity is not printed. If neither true nor false is entered on the line, true is assumed. Optionally, t [ rue ] or f [ alse ].
Line 15: -ionic_strength [( True or False )]
-ionic_strength --Prints ionic strength to the selected-output file. Default is false at startup. Optionally, ionic_strength , mu , -io [ nic_strength ], or -mu .
( True or False )--If true , ionic strength is printed to the selected-output file; if false , ionic strength is not printed. If neither true nor false is entered on the line, true is assumed. Optionally, t [ rue ] or f [ alse ].
Line 16: -water [( True or False )]
-water --Prints mass of water to the selected-output file for each calculation. Default is false at startup. Optionally, water or -w [ ater ].
( True or False )--If true , mass of water is printed to the selected-output file; if false , mass of water is not printed. If neither true nor false is entered on the line, true is assumed. Optionally, t [ rue ] or f [ alse ].
Line 17: -charge_balance [( True or False )]
-charge_balance --Prints charge balance of solution (eq, equivalent) to the selected-output file for each calculation. Default is false at startup. Optionally, charge_balance or -c [ harge_balance ].
( True or False )--If true , charge balance is printed to the selected-output file; if false , charge balance is not printed. If neither true nor false is entered on the line, true is assumed. Optionally, t [ rue ] or f [ alse ].
Line 18: -percent_error [( True or False )]
-percent_error
--Prints percent error in charge balance (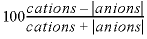 ) to the selected-output file for each calculation. Default is
false
at startup. Optionally,
percent_error
or
-per
[
cent_error
].
) to the selected-output file for each calculation. Default is
false
at startup. Optionally,
percent_error
or
-per
[
cent_error
].
( True or False )--If true , percent error is printed to the selected-output file; if false , percent error is not printed. If neither true nor false is entered on the line, true is assumed. Optionally, t [ rue ] or f [ alse ].
-totals --Identifier allows definition of a list of total concentrations, in molality, that will be written to the selected-output file. Optionally, totals or -t [ otals ].
element list --List of elements, element valence states, exchange sites, or surface sites for which total concentrations will be written. The list may continue on the subsequent line(s) (Line 2a). After each calculation, the concentration (mol/kgw) of each of the selected elements, element valence states, exchange sites, and surface sites will be written to the selected-output file. Elements, valence states, exchange sites, and surface sites are defined in the first column of SOLUTION_MASTER_SPECIES, EXCHANGE_MASTER_SPECIES, or SURFACE_MASTER_SPECIES input. If an element is not defined or is not present in the calculation, its concentration will be printed as 0.
element list --Continuation of a list for -totals of elements, element valence states, exchange sites, or surface sites.
Line 21: -molalities species list
-molalities --Identifier allows definition of a list of species for which concentrations will be written to the selected-output file. Optionally, molalities , mol , or -m [ olalities ].
species list --List of aqueous, exchange, or surface species for which concentrations will be written to the selected-output file. The list may continue on the subsequent line(s). After each calculation, the concentration (mol/kgw) of each species in the list will be written to the selected-output file. Species are defined by SOLUTION_SPECIES, EXCHANGE_SPECIES, or SURFACE_SPECIES input. If a species is not defined or is not present in the calculation, its concentration will be printed as 0.
Line 22: -activities species list
-activities --Identifier allows definition of a list of species for which log of activity will be written to the selected-output file. Optionally, activities or -a [ ctivities ].
species list --List of aqueous, exchange, or surface species for which log of activity will be written to the selected-output file. The list may continue on the subsequent line(s). After each calculation, the log (base 10) of the activity of each of the species will be written to the selected-output file. Species are defined by SOLUTION_SPECIES, EXCHANGE_SPECIES, or SURFACE_SPECIES input. If a species is not defined or is not present in the calculation, its log activity will be printed as -999.999.
Line 23: -equilibrium_phases phase list
-equilibrium_phases --Identifier allows definition of a list of pure phases for which (1) total amounts in the pure-phase assemblage and (2) moles transferred will be written to the selected-output file. Optionally, -e[quilibrium_phases ] or -p [ ure_phases ]. Note the hyphen is required to avoid a conflict with the keyword EQUILIBRIUM_PHASES and its synonyms.
phase list --List of phases for which data will be written to the selected-output file. The list may continue on the subsequent line(s). After each calculation, two values are written to the selected-output file: (1) the moles of each of the phases (defined by EQUILIBRIUM_PHASES), and (2) the moles transferred. Phases are defined by PHASES input. If the phase is not defined or is not present in the pure-phase assemblage, the amounts will be printed as 0.
Line 24: -saturation_indices phase list
-saturation_indices --Identifier allows definition of a list of phases for which saturation indices [or log (base 10) fugacity for gases] will be written to the selected-output file. Optionally, saturation_indices , si , -s [ aturation_indices ], or -s [ i ].
phase list --List of phases for which saturation indices [or log (base 10) partial pressure for gases] will be written to the selected-output file. The list may continue on the subsequent line(s). After each calculation, the saturation index of each of the phases will be written to the selected-output file. Phases are defined by PHASES input. If the phase is not defined or if one or more of its constituent elements is not in solution, the saturation index will be printed as -999.999.
Line 25: -gases gas-component list
-gases --Identifier allows definition of a list of gas components for which the amount in the gas phase (mol) will be written to the selected-output file. Optionally, gases or -g [ ases ].
gas-component list --List of gas components. The list may continue on the subsequent line(s). After each calculation, the moles of each of the selected gas components in the gas phase will be written to the selected-output file. Gas components are defined by PHASES input. If a gas component is not defined or is not present in the gas phase, the amount will be printed as 0. Before the columns for the gas components, the selected-output file will contain the total pressure, total moles of gas components, and the volume of the gas phase. Log partial pressures of any gas, including the components in the gas phase, can be obtained by use of the -saturation_indices identifier.
Line 26: -kinetic_reactants reactant list
-kinetic_reactants --Identifier allows definition of a list of kinetically controlled reactants for which two values are written to the selected-output file: (1) the current moles of the reactant, and (2) the moles transferred of the reactant. Optionally, kin , -k [ inetics ], kinetic_reactants , or -k [ inetic_reactants ]. Note the hyphen is required to avoid a conflict with the keyword KINETICS.
reactant list --List of kinetically controlled reactants. The list may continue on the subsequent line(s). After each calculation, the moles and the moles transferred of each of the kinetically controlled reactants will be written to the selected-output file. Kinetic reactants are identified by the rate name in the KINETICS data block. (The rate name in turn refers to a rate expression defined with RATES data block.) If the kinetic reactant is not defined, the amounts will be printed as 0.
Line 27: -solid_solutions component list
-solid_solutions --Identifier allows definition of a list of solid-solution components for which the moles in a solid solution is written to the selected-output file. Optionally, -so [ lid_solutions ]. Note the hyphen is required to avoid a conflict with the keyword SOLID_SOLUTIONS.
component list --List of solid-solution components. The list may continue on the subsequent line(s). After each calculation, the moles of each solid-solution component in the list will be written to the selected-output file. A solid-solution component is identified by the component name defined in the SOLID_SOLUTIONS data block. (The component names are also phase names that have been defined in the PHASES data block.) If the component is not defined in any of the solid solutions, the amount will be printed as 0.
Line 28: -isotopes isotope ratio list
-isotopes --Identifier selects isotopes for which values are written to the selected-output file. The units of the isotopic values printed to the selected output file are the units of the standard as entered with keyword ISOTOPES (for example, permil or percent modern carbon). Optionally, -is [ otopes ]. Note the hyphen is required to avoid a conflict with the keyword ISOTOPES.
isotope ratio list --List of ratios for isotopes and isotopic species to be written to the selected-output file. The list may continue on the subsequent line(s). After each calculation, the isotope ratios in the list will be written to the selected-output file. Isotope ratios are defined in the ISOTOPE_RATIOS data block and refer to Basic programs defined in CALCULATE_VALUES data blocks. If an isotope ratio is not defined or is absent from solution, the value printed is -9,999.999.
Line 29: -calculate_values calculate values list
-calculate_values --Identifier selects Basic functions for which function values will be written to the selected-output file. The list may continue on the subsequent line(s). After each calculation, the values for functions in the list will be written to the selected-output file. The Basic functions are defined in the CALCULATE_VALUES data block. Optionally, -ca [ lculate_values ]. Note the hyphen is required to avoid a conflict with the keyword CALCULATE_VALUES.
calculate values list --List of names of Basic functions; value calculated by function will be written to the selected-output file. The list may continue on the subsequent line(s).
Line 30: -inverse_modeling [( True or False )]
-inverse_modeling --Prints results of inverse modeling to the selected-output file. For each inverse model, three values are printed for each solution and phase defined in the INVERSE_MODELING data block: the central value of the mixing fraction or mole transfer, and the minimum and maximum of the mixing fraction or mole transfer, which are zero unless -range is specified in the INVERSE_MODELING data block. Default is true at startup. Optionally, inverse or -i [ nverse_modeling ]. Note the hyphen is required to avoid a conflict with the keyword INVERSE_MODELING .
( True or False )--If true , results of inverse modeling are printed to the selected-output file; if false , results of inverse modeling are not printed. If neither true nor false is entered on the line, true is assumed. Optionally, t [ rue ] or f [ alse ].
Line 31: -active [(True or False)]
-active--Prints results to the selected-output file defined for this SELECTED_OUTPUT data block..Default is true at startup.
(True or False)--If true, selected-output results are written to file; if false, selected-output results are not printed. If neither true nor false is entered on the line, true is assumed. Optionally, t[rue] or f[alse].
Line 32: -user_punch [(True or False)]
-user_punch--Prints USER_PUNCH n results to the selected-output file defined for this SELECTED_OUTPUT data block when n is the number of this SELECTED_OUTPUT data block. Default is true at startup. Note the hyphen is required to avoid a conflict with the keyword USER_PUNCH.
(True or False)--If true, USER_PUNCH n results are printed to the selected-output file; if false, USER_PUNCH n results are printed are not printed. If neither true nor false is entered on the line, true is assumed. Optionally, t[rue] or f[alse].