For Each Simulation
|
Data Set 1
|
MAXRIP MAXPOLY IRIPCB IRIPCB1
MAXRIP is the maximum number of riparian cells.
|
MAXPOLY is the maximum number of polygons in a cell.
|
IRIPCB is a cell-by-cell flow flag and unit number.
• IRIPCB>0, unit number to which the total evapotranspiration rate for each cell will be written when SAVE BUDGET or a non-zero value of ICBCFL is specified in output control.
•IRIPCB=0, cell-by-cell terms not written.
•IRIPCB<0, the ET rate for each plant functional subgroup will be written to the LIST file by cell when SAVE BUDGET or a non-zero value of ICBCFL is specified in output control. |
IRIPCB1 is a flag and unit number.
•IRIPCB1>0, unit number to which the location, land-surface elevation, and transpiration rates for each plant functional subgroup and cell are saved.
•IRIPCB1≤0, the values are not saved. |
|
|
Data Set 2
|
MAXTS MXSEG
MAXTS is the total number of plant functional subgroups. At present, MAXTS should less than or equal to 20.
|
MXSEG is the maximum number of segments in any plant functional subgroup for the interpolation of transpiration canopy flux as a function of hydraulic head.
|
|
|
Items 3, 4, and 5 are read for each plant functional subgroup.
|
Data Set 3
|
RIPNM Sxd Ard Rmax Rsxd NuSeg
RIPNM are the plant functional subgroup names, such as:
Obligate wetland: low, medium or high density.
Shallow-rooted riparian: low, medium, or high density.
Deep-rooted riparian: small, medium, or large size.
Transitional-riparian: small, medium, or large size.
RIPNM is string of up to 24 characters.
|
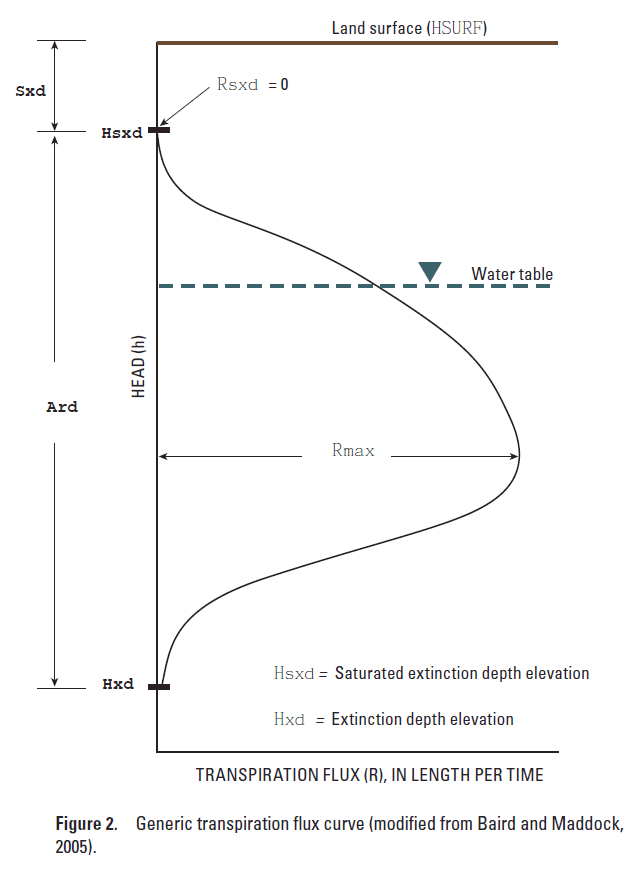
Sxd is the saturated extinction depth with respect to the surface elevation, (L). (A negative depth is above land surface).
|
Ard is the active root depth, (L).

|
Rmax is the maximum transpiration or evaporation flux, (L/t).

|
Rsxd is the transpiration canopy flux at the saturated extinction depth or the maximum evaporation rate, (L/t).

|
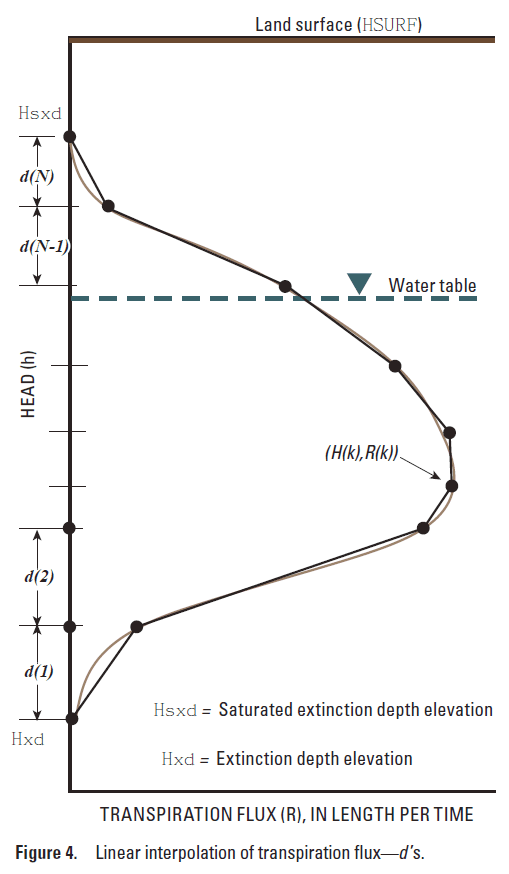
NuSeg is the number of active segments to perform a linear interpolation for a plant functional subgroup.
|
|
|
Data Set 4
|
fdh(1) fdh(2) ... fdh(NuSeg)
fdh is the dimensionless active root depth segment (eqn. 5.1, dimensionless).
|
|
Data Set 5
|
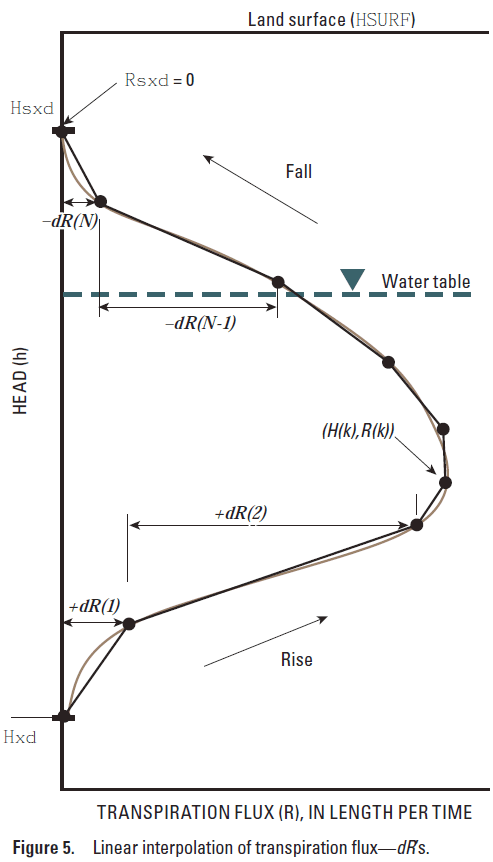
fdR(1) fdR(2) ... fdR(NuSeg)
fdR is the dimensionless flux segment (eqn. 5.2, dimensionless).
|
|
For Each Stress Period
|
Data Set 6
|
ITMP is a flag or the number of riparian cells.
•≥ 0, number of riparian cells active during the current stress period.
•< 0, same riparian cells active during last stress period will be active during the current stress period. |
|
Repeat 7 and 8 until all riaparian cells have been read.
|
The following data are read for each riparian cell.
|
Data Set 7
|
Layer Row Column NPOLY
Layer is the layer number of the riparian cell.
|
Row is the row number of the riparian cell.
|
Column is the column number of the riparian cell.
|
NPOLY is the number of polygons in a cell.
|
|
|
And for each polygon within the riparian cell, read data set 8
|
Data Set 8
|
HSURF fCov(1) ... fCov(MAXTS)
HSURF is the land-surface elevation of a polygon in a riparian cell.
|
fCov is the fraction of land coverage of a plant functional subgroup for a polygon within a cell; for each polygon, there may be multiple plant functional subgroups.
|
|
|



