Explanation 1
Line 0:
PITZER
PITZER
is the keyword for the data block. No other data are input on the keyword line.
Line 1:
-macinnes
[(
True
or
False
)]
-macinnes
--Identifier determines whether activity coefficients printed in the output are scaled by the MacInnes assumption that the activity coefficients of Cl
-
and K
+
are equal (see Plummer and others, 1988). Default is
true
at startup. Optionally,
macinnes
or
-m
[
acinnes
].
(
True
or
False
)--A value of
true
indicates that activity coefficients printed in the output are scaled by the MacInnes assumption;
false
indicates that the activity coefficients printed in the output are unscaled. If neither
true
nor
false
is entered on the line,
true
is assumed. Optionally,
t
[
rue
] or
f
[
alse
].
Line 2:
-use_etheta
[(
True
or
False
)]
-use_etheta
--Identifier determines whether nonsymmetric mixing coefficients are used in the calculations (see Plummer and others, 1988, for a description of the coefficients 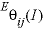 and
and 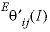 ). Default is
true
at startup. Optionally,
use_etheta
or
-u
[
se_etheta
].
). Default is
true
at startup. Optionally,
use_etheta
or
-u
[
se_etheta
].
(
True
or
False
)--A value of
true
indicates that the nonsymmetric mixing coefficients will be calculated;
false
indicates that the nonsymmetric mixing coefficients will be set to 1.0.If neither
true
nor
false
is entered on the line,
true
is assumed. Optionally,
t
[
rue
] or
f
[
alse
].
Line 3:
-redox
[(
True
or
False
)]
-redox
--Identifier determines whether a redox-related equation is included in the calculations. This option is useful only if using a database that contains at least one redox couple. Note that the pitzer.dat database does not include any redox couples and thus cannot simulate any redox reactions. Default is
false
at startup. Optionally,
redox
or
-r
[
edox
].
(
True
or
False
)--A value of
true
indicates that a redox-related equation will be included;
false
indicates that a redox-related equation is not included. If neither
true
nor
false
is entered on the line,
true
is assumed. Optionally,
t
[
rue
] or
f
[
alse
].
Explanation 2
Line 0:
PITZER
PITZER
is the keyword for the data block. No other data are input on the keyword line.
Line 1:
-B0
-B0
--Identifier begins a block of data that defines 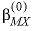 cation-anion interaction parameters for the Pitzer aqueous model (see Plummer and others, 1988).
cation-anion interaction parameters for the Pitzer aqueous model (see Plummer and others, 1988).
Line 2:
cation anion A
0
,
A
1
,
A
2
,
A
3
,
A
4
,
A
5
cation anion
--A cation-anion pair.
A
0
,
A
1
,
A
2
,
A
3
,
A
4
,
A
5
--Coefficients for the temperature dependence of the Pitzer parameter. The expression for a Pitzer parameter is as follows: 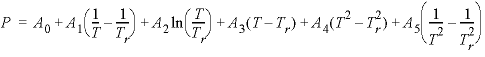 , where
P
is the parameter,
T
is the temperature in kelvin,
T
r
is the reference temperature (298.15 K), and ln is the natural log. If less than six parameters are defined, the undefined parameters are assumed to be zero.
, where
P
is the parameter,
T
is the temperature in kelvin,
T
r
is the reference temperature (298.15 K), and ln is the natural log. If less than six parameters are defined, the undefined parameters are assumed to be zero.
Line 3:
-B1
-B1
--Identifier begins a block of data that defines 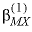 cation-anion interaction parameters for the Pitzer aqueous model (see Plummer and others, 1988).
cation-anion interaction parameters for the Pitzer aqueous model (see Plummer and others, 1988).
Line 4:
-B2
-B2
--Identifier begins a block of data that defines 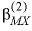 cation-anion interaction parameters for the Pitzer aqueous model (see Plummer and others, 1988).
cation-anion interaction parameters for the Pitzer aqueous model (see Plummer and others, 1988).
Line 5:
-C0
-C0
--Identifier begins a block of data that defines 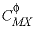 cation-anion interaction parameters for the Pitzer aqueous model (see Plummer and others, 1988).
cation-anion interaction parameters for the Pitzer aqueous model (see Plummer and others, 1988).
Line 6:
-THETA
-THETA
--Identifier begins a block of data that defines 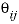 cation-cation and anion-anion interaction parameters for the Pitzer aqueous model (see Plummer and others, 1988).
cation-cation and anion-anion interaction parameters for the Pitzer aqueous model (see Plummer and others, 1988).
Line 7: (
cation cation
or
anion anion
)
A
0
,
A
1
,
A
2
,
A
3
,
A
4
,
A
5
(
cation cation
or
anion anion
)--A cation-cation pair of ions or an anion-anion pair of ions.
A
0
,
A
1
,
A
2
,
A
3
,
A
4
,
A
5
--Coefficients for the temperature dependence of the Pitzer parameter (see Line 2).
Line 8:
-LAMBDA
-LAMBDA
--Identifier begins a block of data that defines 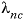 neutral-cation or
neutral-cation or 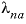 neutral-anion interaction parameters for the Pitzer aqueous model (see Plummer and others, 1988).
neutral-anion interaction parameters for the Pitzer aqueous model (see Plummer and others, 1988).
Line 9: (
neutral cation
or
neutral anion
)
A
0
,
A
1
,
A
2
,
A
3
,
A
4
,
A
5
(
neutral cation
or
neutral anion
)--A neutral-cation pair of species or a neutral-anion pair of species.
A
0
,
A
1
,
A
2
,
A
3
,
A
4
,
A
5
--Coefficients for the temperature dependence of the Pitzer parameter (see Line 2).
Line 10:
-PSI
-PSI
--Identifier begins a block of data that defines 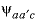 anion-anion-cation (where
a
and
anion-anion-cation (where
a
and 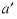 are dissimilar) or
are dissimilar) or 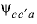 cation-cation-anion (where
c
and
cation-cation-anion (where
c
and 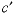 are dissimilar) interaction parameters for the Pitzer aqueous model (see Plummer and others, 1988).
are dissimilar) interaction parameters for the Pitzer aqueous model (see Plummer and others, 1988).
Line 11: (
cation cation anion
or
anion anion cation
)
A
0
,
A
1
,
A
2
,
A
3
,
A
4
,
A
5
(
cation cation anion
or
anion anion cation
)--A cation-cation-anion triple of ions or an anion-anion-cation triple of ions.
A
0
,
A
1
,
A
2
,
A
3
,
A
4
,
A
5
--Coefficients for the temperature dependence of the Pitzer parameter (see Line 2).
Line 12:
-ZETA
-ZETA
--Identifier begins a block of data that defines neutral-cation-anion (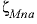 ,
, 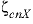 , or
, or 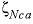 where
M
and
c
represent cations, and
a
and
X
represent anions, and
N
and
n
represent neutral species) interaction parameters for the Pitzer aqueous model (see Clegg and Whitfield, 1991; Clegg and Whitfield, 1995, p. 2404 corrects the coefficient of the zeta term in the 1991 paper).
where
M
and
c
represent cations, and
a
and
X
represent anions, and
N
and
n
represent neutral species) interaction parameters for the Pitzer aqueous model (see Clegg and Whitfield, 1991; Clegg and Whitfield, 1995, p. 2404 corrects the coefficient of the zeta term in the 1991 paper).
Line 13:
neutral cation anion A
0
,
A
1
,
A
2
,
A
3
,
A
4
,
A
5
neutral cation anion
--A neutral-cation-anion triple of species.
A
0
,
A
1
,
A
2
,
A
3
,
A
4
,
A
5
--Coefficients for the temperature dependence of the Pitzer parameter (see Line 2).
Line 14:
-MU
-MU
--Identifier begins a block of data that defines 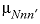 neutral-neutral-neutral (where
n and
neutral-neutral-neutral (where
n and 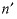 represent dissimilar neutral species) interaction parameters for the Pitzer aqueous model (see Clegg and Whitfield, 1991).
represent dissimilar neutral species) interaction parameters for the Pitzer aqueous model (see Clegg and Whitfield, 1991).
Line 15:
neutral neutral neutral A
0
,
A
1
,
A
2
,
A
3
,
A
4
,
A
5
neutral neutral neutral
--A neutral-neutral-neutral triple of species.
A
0
,
A
1
,
A
2
,
A
3
,
A
4
,
A
5
--Coefficients for the temperature dependence of the Pitzer parameter (see Line 2).
Line 16:
-ETA
-ETA
--Identifier begins a block of data that defines neutral-anion-anion and neutral-cation-cation (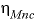 ,
, 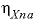 ,
, 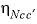 , or
, or 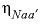 , where
M
and
c
represent cations, and
a
and
X
represent anions, and
N
and
n
represent neutral species, and prime indicated dissimilar species) interaction parameters for the Pitzer aqueous model (see Clegg and Whitfield, 1991; Clegg and Whitfield, 1995, p. 2404 corrects the coefficient of the eta term in the 1991 paper).
, where
M
and
c
represent cations, and
a
and
X
represent anions, and
N
and
n
represent neutral species, and prime indicated dissimilar species) interaction parameters for the Pitzer aqueous model (see Clegg and Whitfield, 1991; Clegg and Whitfield, 1995, p. 2404 corrects the coefficient of the eta term in the 1991 paper).
Line 17: (
neutral cation cation
or
neutral anion anion
)
A
0
,
A
1
,
A
2
,
A
3
,
A
4
,
A
5
(
neutral cation cation
or
neutral anion anion
)--A neutral-cation-cation or neutral-anion-anion triple of species.
A
0
,
A
1
,
A
2
,
A
3
,
A
4
,
A
5
--Coefficients for the temperature dependence of the Pitzer parameter (see Line 2).
Line 18:
-alphas
-alphas
--Identifier begins a block of data that defines alpha parameters for the Pitzer aqueous model that override default values for specific cation-anion pairs. For any electrolyte containing a monovalent ion, a single 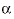 parameter with the default value of 2.0 is used in the calculation of
parameter with the default value of 2.0 is used in the calculation of 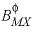 ,
, 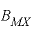 , and
, and 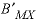 . For electrolytes containing two polyvalent ions, two parameters,
. For electrolytes containing two polyvalent ions, two parameters, 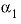 and
and 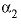 , are used in the calculation of
, are used in the calculation of 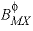 ,
, 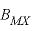 , and
, and 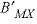 . For 2-2 electrolytes, the defaults are
. For 2-2 electrolytes, the defaults are 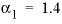 and
and 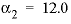 . For 3-2 and 4-2 electrolytes, the defaults are
. For 3-2 and 4-2 electrolytes, the defaults are 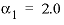 and
and 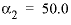 (see Plummer and others, 1988).
(see Plummer and others, 1988).
Line 19:
cation anion
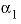
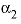
cation anion
--A cation-anion pair.
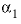 --Value of the
--Value of the 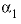 parameter. For electrolytes with at least one monovalent ion,
parameter. For electrolytes with at least one monovalent ion, 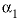 is interpreted as
is interpreted as 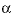 .
.
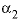 --Value of the
--Value of the 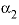 parameter. For electrolytes with at least one monovalent ion,
parameter. For electrolytes with at least one monovalent ion, 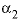 is not used.
is not used.
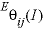 and
and 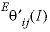 ). Default is
true
at startup. Optionally,
use_etheta
or
-u
[
se_etheta
].
). Default is
true
at startup. Optionally,
use_etheta
or
-u
[
se_etheta
].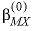 cation-anion interaction parameters for the Pitzer aqueous model (see Plummer and others, 1988).
cation-anion interaction parameters for the Pitzer aqueous model (see Plummer and others, 1988).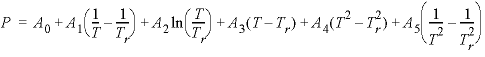 , where
P
is the parameter,
T
is the temperature in kelvin,
T
r
is the reference temperature (298.15 K), and ln is the natural log. If less than six parameters are defined, the undefined parameters are assumed to be zero.
, where
P
is the parameter,
T
is the temperature in kelvin,
T
r
is the reference temperature (298.15 K), and ln is the natural log. If less than six parameters are defined, the undefined parameters are assumed to be zero. 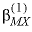 cation-anion interaction parameters for the Pitzer aqueous model (see Plummer and others, 1988).
cation-anion interaction parameters for the Pitzer aqueous model (see Plummer and others, 1988).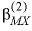 cation-anion interaction parameters for the Pitzer aqueous model (see Plummer and others, 1988).
cation-anion interaction parameters for the Pitzer aqueous model (see Plummer and others, 1988).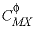 cation-anion interaction parameters for the Pitzer aqueous model (see Plummer and others, 1988).
cation-anion interaction parameters for the Pitzer aqueous model (see Plummer and others, 1988).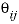 cation-cation and anion-anion interaction parameters for the Pitzer aqueous model (see Plummer and others, 1988).
cation-cation and anion-anion interaction parameters for the Pitzer aqueous model (see Plummer and others, 1988).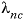 neutral-cation or
neutral-cation or 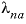 neutral-anion interaction parameters for the Pitzer aqueous model (see Plummer and others, 1988).
neutral-anion interaction parameters for the Pitzer aqueous model (see Plummer and others, 1988).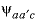 anion-anion-cation (where
a
and
anion-anion-cation (where
a
and 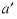 are dissimilar) or
are dissimilar) or 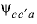 cation-cation-anion (where
c
and
cation-cation-anion (where
c
and 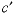 are dissimilar) interaction parameters for the Pitzer aqueous model (see Plummer and others, 1988).
are dissimilar) interaction parameters for the Pitzer aqueous model (see Plummer and others, 1988).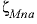 ,
, 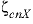 , or
, or 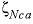 where
M
and
c
represent cations, and
a
and
X
represent anions, and
N
and
n
represent neutral species) interaction parameters for the Pitzer aqueous model (see Clegg and Whitfield, 1991; Clegg and Whitfield, 1995, p. 2404 corrects the coefficient of the zeta term in the 1991 paper).
where
M
and
c
represent cations, and
a
and
X
represent anions, and
N
and
n
represent neutral species) interaction parameters for the Pitzer aqueous model (see Clegg and Whitfield, 1991; Clegg and Whitfield, 1995, p. 2404 corrects the coefficient of the zeta term in the 1991 paper).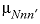 neutral-neutral-neutral (where
n and
neutral-neutral-neutral (where
n and 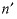 represent dissimilar neutral species) interaction parameters for the Pitzer aqueous model (see Clegg and Whitfield, 1991).
represent dissimilar neutral species) interaction parameters for the Pitzer aqueous model (see Clegg and Whitfield, 1991).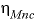 ,
, 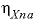 ,
, 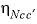 , or
, or 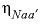 , where
M
and
c
represent cations, and
a
and
X
represent anions, and
N
and
n
represent neutral species, and prime indicated dissimilar species) interaction parameters for the Pitzer aqueous model (see Clegg and Whitfield, 1991; Clegg and Whitfield, 1995, p. 2404 corrects the coefficient of the eta term in the 1991 paper).
, where
M
and
c
represent cations, and
a
and
X
represent anions, and
N
and
n
represent neutral species, and prime indicated dissimilar species) interaction parameters for the Pitzer aqueous model (see Clegg and Whitfield, 1991; Clegg and Whitfield, 1995, p. 2404 corrects the coefficient of the eta term in the 1991 paper).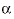 parameter with the default value of 2.0 is used in the calculation of
parameter with the default value of 2.0 is used in the calculation of 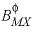 ,
, 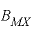 , and
, and 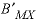 . For electrolytes containing two polyvalent ions, two parameters,
. For electrolytes containing two polyvalent ions, two parameters, 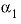 and
and 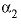 , are used in the calculation of
, are used in the calculation of 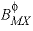 ,
, 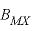 , and
, and 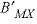 . For 2-2 electrolytes, the defaults are
. For 2-2 electrolytes, the defaults are 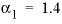 and
and 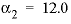 . For 3-2 and 4-2 electrolytes, the defaults are
. For 3-2 and 4-2 electrolytes, the defaults are 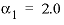 and
and 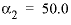 (see Plummer and others, 1988).
(see Plummer and others, 1988).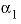
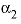
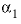 --Value of the
--Value of the 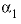 parameter. For electrolytes with at least one monovalent ion,
parameter. For electrolytes with at least one monovalent ion, 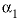 is interpreted as
is interpreted as 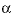 .
. 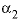 --Value of the
--Value of the 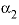 parameter. For electrolytes with at least one monovalent ion,
parameter. For electrolytes with at least one monovalent ion, 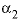 is not used.
is not used.