Use Cases
Catchment characteristics ScienceBase collection
Keywords: model parameters; basin characterization; site selection
Domain: Climate; Geology; Hydrogeology; Hydrology; Infrastructure; Land Cover; Soils; Stream Characteristics; Topography; Water Use
Language: R
Description:
Multiple variables of natural and anthropogenic landscape features linked to NHDPlus v2 (stream segments, catchments, and upstream watersheds) within the conterminous United States. These data allow researchers to acquire landscape information for both catchments and full upstream watersheds of specific stream reaches anywhere in the within the conterminous United States without having to perform specialized geospatial processing.
Linked Catalog Datasets:
Linked Catalog Tools:
Purpose
The USGS has over 4,000 natural and anthropogenic landscape characteristics linked to NHDPlus Version 2.1’s (NHDPlusV2) approximately 2.7 million stream segments, their associated catchments, and their upstream watersheds within the conterminous United States. ( https://doi.org/10.5066/F7765D7V ) The nhdplusTools package is a growing collection of R tools that enables researchers to gather landscape information for both flowline catchments and/or entire upstream watersheds without the need for complex geospatial processing or downloading huge amounts of data. This script demonstrates using the nhdplusTools version 0.7.1 to download (subsets of) NHDPlusV2 spatial data and their associated landscape characteristics from a cloud data store and mapping the results.
Description
This use case demonstrates how to access upstream characteristics using functions included in nhdplusTools version 0.7.1. for both a single flowline catchment and multiple catchments for a USGS streamgage and mapping the results:
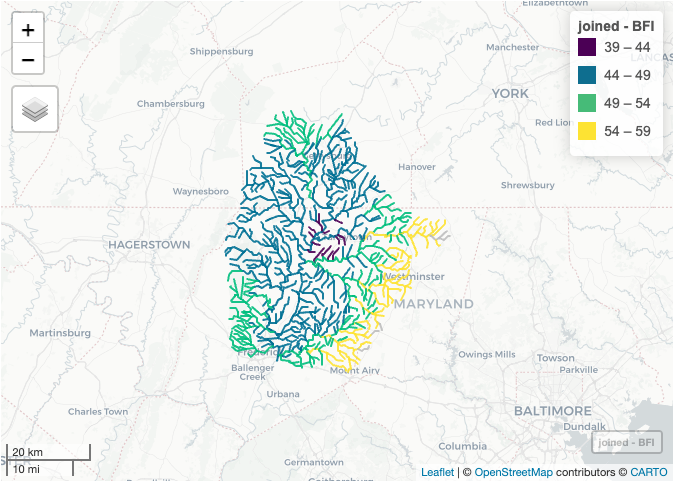
- Base Flow Index (BFI, https://www.sciencebase.gov/catalog/item/5669a8e3e4b08895842a1d4f )
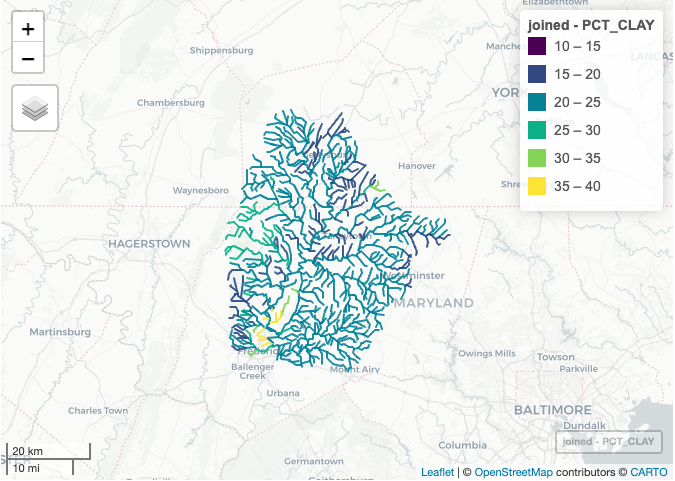
- Percent clay (PCT_CLAY, https://www.sciencebase.gov/catalog/item/5728dd46e4b0b13d3918a9a7 )
See the metadata in each link above for details on the routing metrics and description of the variables. Both examples, access total upstream routing data, which represents full upstream watershed metrics for a flowline catchment.
Required package nhdplusTools
nhdplusTools version 0.7.1: R package of a growing set of tools for manipulation of hydrographic data using the NHDPlus data model. ( https://doi-usgs.github.io/nhdplusTools/ )
## Getting Started
# Install nhdplusTools version 0.7.1 if not installed
install.packages("remotes")
remotes::install_github("DOI-USGS/nhdplusTools")
Install libraries for use in this demo
See https://doi-usgs.github.io/nhdplusTools/index.html for details on each nhdplusTools function and their options
library(nhdplusTools)
library(dplyr)
library(sf)
library(mapview)
Get BFI value for one COMID
## Get metadata for BFI to ascertain the name of the variable of interest.
glimpse(get_characteristics_metadata(search = "BFI"))
## Access the data value for one NHDPlus version2 COMID and save it into a dataframe
df <- get_catchment_characteristics("TOT_BFI", c(5329343))
df
Get BFI values for each flowline upstream of a USGS streamgage
# Navigate the network upstream of USGS gage, 0164300 Monocacy River in MD,
# and obtain upstream NHDPlus version2 flowlines. Save to a dataframe.
nhd <- navigate_network(list(featureSource = "nwissite", featureID = "USGS-01643000"), "UT", output = "flowlines", trim_start = TRUE, distance_km = 100)
comids <- nhd$comid
# Using the list of COMIDs, obtain BFI values for all upstream flowlines for
# 0164300 Monocacy River watershed in MD
# Perform some summary statistics to use the range of values for mapping purposes
nhd_bfi <- get_catchment_characteristics("TOT_BFI", comids, reference_fabric="nhdplusv2")
summary(nhd_bfi)
# Join the BFI data to the spatial layer of flowlines
joined <- merge(nhd, nhd_bfi, characteristic_value, by.x = "comid", by.y = "comid")
## Change column name to something meaningful
colnames(joined)[colnames(joined) == "characteristic_value"] ="BFI"
# Map the data
mapview(joined, zcol = "BFI", at = seq(39, 61, 5), legend = TRUE)
Get percent clay values for each flowline upstream of a USGS streamgage
# Get metadata for percent clay to ascertain the name of the variable of interest.
glimpse(get_characteristics_metadata(search = "CLAY"))
# Navigate the network upstream of USGS gage, 0164300 Monocacy River in MD,
# and obtain upstream NHDPlus version2 flowlines. Save to a dataframe.
# Using the list of COMIDs, obtain BFI values for all upstream flowlines for
# 0164300 Monocacy River watershed in MD
# Perform some summary statistics to use the range of values for mapping purposes
nhd_clay <- get_catchment_characteristics("TOT_CLAYAVE", comids, reference_fabric="nhdplusv2")
summary(nhd_clay)
## Join the percent clay data to the spatial layer of flowlines
joined <- merge(nhd, nhd_clay, characteristic_value, by.x = "comid", by.y = "comid")
# Change column name to something meaningful
colnames(joined)[colnames(joined) == "characteristic_value"] ="PCT_CLAY"
# Map the data
mapview(joined, zcol = "PCT_CLAY", at = seq(10, 40, 5), legend = TRUE)
References
Wieczorek, M.E., Jackson, S.E., and Schwarz, G.E., 2018, Select Attributes for NHDPlus Version 2.1 Reach Catchments and Modified Network Routed Upstream Watersheds for the Conterminous United States (ver. 4.0, August 2023): U.S. Geological Survey data release, https://doi.org/10.5066/F7765D7V .
Blodgett, D., Johnson, J.M., 2022, nhdplusTools: Tools for Accessing and Working with the NHDPlus, https://doi.org/10.5066/P97AS8JD .

