# Common python packages
import xarray as xr
import hvplot.xarray
import hvplot.pandas
import hvplot.dask
import intake
import warnings
import intake_xarray
import datetime
import holoviews as hv
import numpy as np
import pandas as pd
import geopandas as gpd
# HyRiver packages
from pynhd import NLDI, WaterData
# GDPTools packages
from gdptools import AggGen, UserCatData, WeightGen
hv.extension("bokeh")
warnings.filterwarnings('ignore')Use Cases
gdptools-conus404
Keywords: CONUS-404; gdptools; spatial-interpolation
Domain: Climate; Hydrology
Language: Python
Description:
This notebook demonstrates the use of GDPTools for spatially interpolating (Aggregation) Conus404 data to a set of HUC12s in the Delaware River Basin.
GDPTools: Conus404-daily Aggregation
This notebook demonstrates the use of GDPTools for spatially interpolating (Aggregation) Conus404 data to a set of HUC12s in the Delaware River Basin.
Summary
This use-case demonstrates a common spatial interpolation pattern in hydrologic sciences. Interpolating gridded data, in this case the Conus404 dataset, to Polygons representing basins, such as HUC12s. GDPTools is a pyhton package for Geospatial Interpolation, developed by the USGS NHGF project. We use GDPTools to generate a weights file for area-weighted spatial interpolation, interpolating the time-dependend gridded data to the HUC12s. The Conus404 data release describes Conus404 as “… a unique, high-resolution hydro-climate dataset appropriate for forcing hydrological models and conducting meteorological analysis over the contiguous United States”. This use-case uses the HyTest intake catalog which provides a convenient method to access the Conus404 data.
Open Source python Tools
- gdptools: GeoDataProcessing Tools (GDPTools) is a python package for geospatial interpolation
- HyTest intake catalog
- NHGF data service clients:
Imports
GDPTools version
Note this use-case is currently using the following version of GDPTools. GDPTools is in an active state of development, and if the viewer of this use-case would like to use GDPTools for their work, it would be worth checking if you have the most up-to-date version.
!conda list gdptools# packages in environment at /opt/homebrew/Caskroom/miniforge/base/envs/gdptools-examples:
#
# Name Version Build Channel
gdptools 0.2.7 pyhd8ed1ab_0 conda-forgeHyRiver Packages
Here we use two HyRiver clients, NLDI and WaterData1 to: - Define the upstream basin at a USGS gage towards the downstream reach of the Delaware River - Use the basin geometry to extract HUC21s within the geometry.
# USGS gage 01482100 Delaware River at Del Mem Bridge at Wilmington De
gage_id = '01482100'
nldi = NLDI()
del_basins = nldi.get_basins(gage_id)
huc12_basins = WaterData('wbd12').bygeom(del_basins.geometry[0])View the content of the resulting GeoDataFrame
huc12_basins.head()| geometry | objectid | tnmid | metasourceid | sourcedatadesc | sourceoriginator | sourcefeatureid | loaddate | gnis_id | areaacres | areasqkm | states | huc12 | name | hutype | humod | tohuc | noncontributingareaacres | noncontributingareasqkm | globalid | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | MULTIPOLYGON (((-75.69696 39.63149, -75.69653 ... | 20165 | {0F437B7A-8CC7-4910-87AE-470DF920F088} | None | None | None | None | 2013-01-18T07:08:10Z | None | 9569.63 | 38.73 | DE,MD | 020402050501 | Muddy Run | S | NM | 020402050502 | 0 | 0 | {9161E1FC-E29C-11E2-8094-0021280458E6} |
| 1 | MULTIPOLYGON (((-75.74141 39.57760, -75.74140 ... | 20627 | {9FDAEFFF-4386-409F-BAB5-C2D1D450DDEF} | {398C29E1-B69C-4748-A790-3729C5B5D492} | None | None | None | 2017-10-17T09:14:29Z | None | 24274.41 | 98.24 | DE,MD | 020600020204 | Long Creek-Back Creek | S | NM | 020600020205 | 0 | 0 | {9192B677-E29C-11E2-8094-0021280458E6} |
| 2 | MULTIPOLYGON (((-75.75512 39.59106, -75.75537 ... | 20628 | {1B20CDFC-A859-4937-BA8B-3DD93DF90631} | {398C29E1-B69C-4748-A790-3729C5B5D492} | None | None | None | 2017-10-17T09:14:29Z | None | 18380.59 | 74.38 | DE,MD | 020600020205 | Perch Creek-Elk River | S | NM | 020600020207 | 0 | 0 | {9192D4A6-E29C-11E2-8094-0021280458E6} |
| 3 | MULTIPOLYGON (((-75.88358 39.78914, -75.88322 ... | 20650 | {21BC2680-D2E8-4F6F-8951-0E42445BB8D8} | None | None | None | None | 2013-01-18T07:08:10Z | None | 30542.71 | 123.60 | DE,MD,PA | 020600020203 | Big Elk Creek | S | NM | 020600020205 | 0 | 0 | {919549F9-E29C-11E2-8094-0021280458E6} |
| 4 | MULTIPOLYGON (((-75.43455 39.78975, -75.43523 ... | 20651 | {A462BFFC-6E58-472A-9996-D8EE655030A4} | {ED602145-9201-4827-9CE1-05D252484579} | None | None | None | 2017-10-03T20:10:51Z | None | 265274.96 | 1073.53 | DE,NJ | 020402040000 | Delaware River-Delaware Bay | W | NM | 020403030601 | 0 | 0 | {9195629C-E29C-11E2-8094-0021280458E6} |
Note the crs associate with the GeoDataFrame
This value will be used later.
huc12_basins.crs<Geographic 2D CRS: EPSG:4326>
Name: WGS 84
Axis Info [ellipsoidal]:
- Lat[north]: Geodetic latitude (degree)
- Lon[east]: Geodetic longitude (degree)
Area of Use:
- name: World.
- bounds: (-180.0, -90.0, 180.0, 90.0)
Datum: World Geodetic System 1984 ensemble
- Ellipsoid: WGS 84
- Prime Meridian: GreenwichReduce the context of the GeoDataFrame to only the columns necessary for the analysis
GDPTools requires only the column used to identify the basins and the geometry. To reduce the memory overhead of the calculations below, it’s a good practice to remove unneccessary columns. First we list the columns and then we keep only huc12 and geometry.
print(huc12_basins.columns)
working_gdf = huc12_basins[["huc12", "geometry"]]Index(['geometry', 'objectid', 'tnmid', 'metasourceid', 'sourcedatadesc',
'sourceoriginator', 'sourcefeatureid', 'loaddate', 'gnis_id',
'areaacres', 'areasqkm', 'states', 'huc12', 'name', 'hutype', 'humod',
'tohuc', 'noncontributingareaacres', 'noncontributingareasqkm',
'globalid'],
dtype='object')Plot the resulting GeoDataFrame
#| vscode: {languageId: python}
from holoviews.element.tiles import OSM
drb = working_gdf.hvplot(
geo=True, coastline='50m', alpha=0.2, frame_width=300,
xlabel="longitude", ylabel="latitude", clabel="monthly water evaporation (mm)",
title="Delaware River HUC12 basins", xlim=(-76.8, -73.8), aspect='equal'
)
drb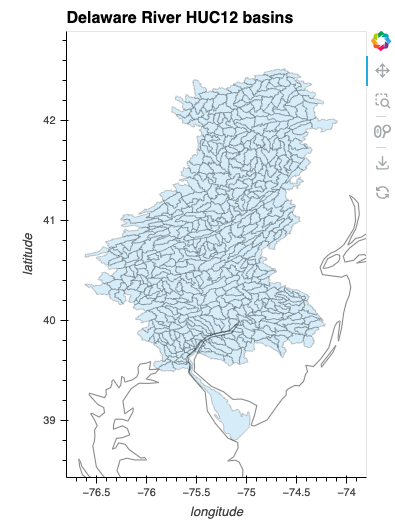
HyTest Intake Catalog and Conus404
Further examples of using the HyTest catalog can be found here. In the following cells we step into the catalog to retrieve the conus404-daily-osn. The data with “-osn” extensions can be accessed without authentication.
Open the catalog
# open the hytest data intake catalog
hytest_cat = intake.open_catalog("https://raw.githubusercontent.com/hytest-org/hytest/main/dataset_catalog/hytest_intake_catalog.yml")
list(hytest_cat)['conus404-catalog',
'conus404-drb-eval-tutorial-catalog',
'nhm-v1.0-daymet-catalog',
'nhm-v1.1-c404-bc-catalog',
'nhm-v1.1-gridmet-catalog',
'trends-and-drivers-catalog',
'nhm-prms-v1.1-gridmet-format-testing-catalog',
'nwis-streamflow-usgs-gages-onprem',
'nwis-streamflow-usgs-gages-osn',
'nwm21-streamflow-usgs-gages-onprem',
'nwm21-streamflow-usgs-gages-osn',
'nwm21-streamflow-cloud',
'geofabric_v1_1-zip-osn',
'geofabric_v1_1_POIs_v1_1-osn',
'geofabric_v1_1_TBtoGFv1_POIs-osn',
'geofabric_v1_1_nhru_v1_1-osn',
'geofabric_v1_1_nhru_v1_1_simp-osn',
'geofabric_v1_1_nsegment_v1_1-osn',
'gages2_nndar-osn',
'wbd-zip-osn',
'huc12-geoparquet-osn',
'huc12-gpkg-osn',
'nwm21-scores',
'lcmap-cloud',
'rechunking-tutorial-osn',
'pointsample-tutorial-sites-osn',
'pointsample-tutorial-output-osn']Open the conus404 catalog
# open the conus404 sub-catalog
cat = hytest_cat['conus404-catalog']
list(cat)['conus404-hourly-onprem',
'conus404-hourly-cloud',
'conus404-hourly-osn',
'conus404-daily-diagnostic-onprem',
'conus404-daily-diagnostic-cloud',
'conus404-daily-diagnostic-osn',
'conus404-daily-onprem',
'conus404-daily-cloud',
'conus404-daily-osn',
'conus404-monthly-onprem',
'conus404-monthly-cloud',
'conus404-monthly-osn',
'conus404-hourly-ba-osn',
'conus404-daily-ba-osn']Open the daily data
## Select the dataset you want to read into your notebook and preview its metadata
dataset = 'conus404-daily-osn'
cat[dataset]conus404-daily-osn:
args:
consolidated: true
storage_options:
anon: true
client_kwargs:
endpoint_url: https://usgs.osn.mghpcc.org/
requester_pays: false
urlpath: s3://hytest/conus404/conus404_daily.zarr
description: CONUS404 40 years of daily values for subset of model output variables
derived from hourly values. You can work with this data for free in any environment
(there are no egress fees).
driver: intake_xarray.xzarr.ZarrSource
metadata:
catalog_dir: https://raw.githubusercontent.com/hytest-org/hytest/main/dataset_catalog/subcatalogsLoad conus404 daily
print(f"Reading {dataset} metadata...", end='')
ds = cat[dataset].to_dask().metpy.parse_cf()
print("done")Reading conus404-daily-osn metadata...doneView the xarray representation
ds<xarray.Dataset> Size: 9TB
Dimensions: (y: 1015, x: 1367, time: 15707, bottom_top_stag: 51,
bottom_top: 50, soil_layers_stag: 4, x_stag: 1368,
y_stag: 1016, snow_layers_stag: 3, snso_layers_stag: 7)
Coordinates:
lat (y, x) float32 6MB dask.array<chunksize=(350, 350), meta=np.ndarray>
lon (y, x) float32 6MB dask.array<chunksize=(350, 350), meta=np.ndarray>
* time (time) datetime64[ns] 126kB 1979-10-01 ... 2022-10-01
* x (x) float64 11kB -2.732e+06 -2.728e+06 ... 2.732e+06
* y (y) float64 8kB -2.028e+06 -2.024e+06 ... 2.028e+06
metpy_crs object 8B Projection: lambert_conformal_conic
lat_u (y, x_stag) float32 6MB dask.array<chunksize=(350, 175), meta=np.ndarray>
lon_u (y, x_stag) float32 6MB dask.array<chunksize=(350, 175), meta=np.ndarray>
lat_v (y_stag, x) float32 6MB dask.array<chunksize=(175, 350), meta=np.ndarray>
lon_v (y_stag, x) float32 6MB dask.array<chunksize=(175, 350), meta=np.ndarray>
Dimensions without coordinates: bottom_top_stag, bottom_top, soil_layers_stag,
x_stag, y_stag, snow_layers_stag,
snso_layers_stag
Data variables: (12/153)
ACDEWC (time, y, x) float32 87GB dask.array<chunksize=(36, 350, 350), meta=np.ndarray>
ACDRIPR (time, y, x) float32 87GB dask.array<chunksize=(36, 350, 350), meta=np.ndarray>
ACDRIPS (time, y, x) float32 87GB dask.array<chunksize=(36, 350, 350), meta=np.ndarray>
ACECAN (time, y, x) float32 87GB dask.array<chunksize=(36, 350, 350), meta=np.ndarray>
ACEDIR (time, y, x) float32 87GB dask.array<chunksize=(36, 350, 350), meta=np.ndarray>
ACETLSM (time, y, x) float32 87GB dask.array<chunksize=(36, 350, 350), meta=np.ndarray>
... ...
ZNU (bottom_top) float32 200B dask.array<chunksize=(50,), meta=np.ndarray>
ZNW (bottom_top_stag) float32 204B dask.array<chunksize=(51,), meta=np.ndarray>
ZS (soil_layers_stag) float32 16B dask.array<chunksize=(4,), meta=np.ndarray>
ZSNSO (time, snso_layers_stag, y, x) float32 610GB dask.array<chunksize=(36, 7, 350, 350), meta=np.ndarray>
ZWT (time, y, x) float32 87GB dask.array<chunksize=(36, 350, 350), meta=np.ndarray>
crs int64 8B ...
Attributes: (12/148)
AER_ANGEXP_OPT: 1
AER_ANGEXP_VAL: 1.2999999523162842
AER_AOD550_OPT: 1
AER_AOD550_VAL: 0.11999999731779099
AER_ASY_OPT: 1
AER_ASY_VAL: 0.8999999761581421
... ...
WEST-EAST_PATCH_START_STAG: 1
WEST-EAST_PATCH_START_UNSTAG: 1
W_DAMPING: 1
YSU_TOPDOWN_PBLMIX: 0
history: Tue Mar 29 16:35:22 2022: ncrcat -A -vW ...
history_of_appended_files: Tue Mar 29 16:35:22 2022: Appended file ...- crs_wkt :
- PROJCRS["unknown",BASEGEOGCRS["unknown",DATUM["unknown",ELLIPSOID["unknown",6370000,0,LENGTHUNIT["metre",1,ID["EPSG",9001]]]],PRIMEM["Greenwich",0,ANGLEUNIT["degree",0.0174532925199433],ID["EPSG",8901]]],CONVERSION["unknown",METHOD["Lambert Conic Conformal (2SP)",ID["EPSG",9802]],PARAMETER["Latitude of false origin",39.1000061035156,ANGLEUNIT["degree",0.0174532925199433],ID["EPSG",8821]],PARAMETER["Longitude of false origin",-97.9000015258789,ANGLEUNIT["degree",0.0174532925199433],ID["EPSG",8822]],PARAMETER["Latitude of 1st standard parallel",30,ANGLEUNIT["degree",0.0174532925199433],ID["EPSG",8823]],PARAMETER["Latitude of 2nd standard parallel",50,ANGLEUNIT["degree",0.0174532925199433],ID["EPSG",8824]],PARAMETER["Easting at false origin",0,LENGTHUNIT["metre",1],ID["EPSG",8826]],PARAMETER["Northing at false origin",0,LENGTHUNIT["metre",1],ID["EPSG",8827]]],CS[Cartesian,2],AXIS["(E)",east,ORDER[1],LENGTHUNIT["metre",1,ID["EPSG",9001]]],AXIS["(N)",north,ORDER[2],LENGTHUNIT["metre",1,ID["EPSG",9001]]]]
- false_easting :
- 0.0
- false_northing :
- 0.0
- geographic_crs_name :
- unknown
- grid_mapping_name :
- lambert_conformal_conic
- horizontal_datum_name :
- unknown
- inverse_flattening :
- 0.0
- latitude_of_projection_origin :
- 39.100006103515625
- longitude_of_central_meridian :
- -97.9000015258789
- longitude_of_prime_meridian :
- 0.0
- prime_meridian_name :
- Greenwich
- projected_crs_name :
- unknown
- reference_ellipsoid_name :
- unknown
- semi_major_axis :
- 6370000.0
- semi_minor_axis :
- 6370000.0
- standard_parallel :
- [30.0, 50.0]
[1 values with dtype=int64]
- timePandasIndex
PandasIndex(DatetimeIndex(['1979-10-01', '1979-10-02', '1979-10-03', '1979-10-04', '1979-10-05', '1979-10-06', '1979-10-07', '1979-10-08', '1979-10-09', '1979-10-10', ... '2022-09-22', '2022-09-23', '2022-09-24', '2022-09-25', '2022-09-26', '2022-09-27', '2022-09-28', '2022-09-29', '2022-09-30', '2022-10-01'], dtype='datetime64[ns]', name='time', length=15707, freq=None)) - xPandasIndex
PandasIndex(Index([-2732000.0, -2728000.0, -2724000.0, -2720000.0, -2716000.0, -2712000.0, -2708000.0, -2704000.0, -2700000.0, -2696000.0, ... 2696000.0, 2700000.0, 2704000.0, 2708000.0, 2712000.0, 2716000.0, 2720000.0, 2724000.0, 2728000.0, 2732000.0], dtype='float64', name='x', length=1367)) - yPandasIndex
PandasIndex(Index([-2027999.9999999958, -2023999.9999999958, -2019999.9999999958, -2015999.9999999958, -2011999.9999999958, -2007999.9999999958, -2003999.9999999958, -1999999.9999999958, -1995999.9999999958, -1991999.9999999958, ... 1992000.0000000042, 1996000.0000000042, 2000000.0000000042, 2004000.0000000042, 2008000.0000000042, 2012000.0000000042, 2016000.0000000042, 2020000.0000000042, 2024000.0000000042, 2028000.0000000042], dtype='float64', name='y', length=1015))
- AER_ANGEXP_OPT :
- 1
- AER_ANGEXP_VAL :
- 1.2999999523162842
- AER_AOD550_OPT :
- 1
- AER_AOD550_VAL :
- 0.11999999731779099
- AER_ASY_OPT :
- 1
- AER_ASY_VAL :
- 0.8999999761581421
- AER_OPT :
- 1
- AER_SSA_OPT :
- 1
- AER_SSA_VAL :
- 0.8500000238418579
- AER_TYPE :
- 1
- BLDT :
- 0.0
- BL_PBL_PHYSICS :
- 1
- BOTTOM-TOP_GRID_DIMENSION :
- 51
- BOTTOM-TOP_PATCH_END_STAG :
- 51
- BOTTOM-TOP_PATCH_END_UNSTAG :
- 50
- BOTTOM-TOP_PATCH_START_STAG :
- 1
- BOTTOM-TOP_PATCH_START_UNSTAG :
- 1
- BUCKET_J :
- 1000000000.0
- BUCKET_MM :
- 100.0
- CEN_LAT :
- 39.100006103515625
- CEN_LON :
- -97.89999389648438
- CUDT :
- 5.0
- CU_PHYSICS :
- 0
- Contacts :
- CHANGHAI LIU (chliu@ucar.edu), KYOKO IKEDA (kyoko@ucar.edu)
- DAMPCOEF :
- 0.20000000298023224
- DAMP_OPT :
- 3
- DFI_OPT :
- 0
- DIFF_6TH_FACTOR :
- 0.11999999731779099
- DIFF_6TH_OPT :
- 0
- DIFF_OPT :
- 2
- DT :
- 20.0
- DTRAMP_MIN :
- 60.0
- DVEG :
- 9
- DX :
- 4000.0
- DY :
- 4000.0
- Division :
- NCAR/RAL/HAP
- ETAC :
- 0.0
- FEEDBACK :
- 1
- FGDT :
- 2.0
- FileGenerated :
- 20210204
- GFDDA_END_H :
- 999999
- GFDDA_INTERVAL_M :
- 180
- GMT :
- 0.0
- GPH :
- 4.999999873689376e-05
- GRAV_SETTLING :
- 0
- GRIDTYPE :
- C
- GRID_FDDA :
- 2
- GRID_ID :
- 1
- GRID_SFDDA :
- 0
- GT :
- 4.999999873689376e-05
- GUV :
- 4.999999873689376e-05
- GWD_OPT :
- 0
- HYBRID_OPT :
- -1
- HYPSOMETRIC_OPT :
- 2
- ICLOUD :
- 1
- ICLOUD_CU :
- 0
- IF_RAMPING :
- 1
- ISFFLX :
- 1
- ISFTCFLX :
- 0
- ISHALLOW :
- 0
- ISICE :
- 15
- ISLAKE :
- 21
- ISOILWATER :
- 14
- ISURBAN :
- 13
- ISWATER :
- 17
- I_PARENT_START :
- 1
- JULDAY :
- 274
- JULYR :
- 1979
- J_PARENT_START :
- 1
- KHDIF :
- 0.0
- KM_OPT :
- 4
- KVDIF :
- 0.0
- MAP_PROJ :
- 1
- MAP_PROJ_CHAR :
- Lambert Conformal
- MFSHCONV :
- 0
- MMINLU :
- MODIFIED_IGBP_MODIS_NOAH
- MOAD_CEN_LAT :
- 39.100006103515625
- MOIST_ADV_OPT :
- 1
- MP_PHYSICS :
- 8
- NCO :
- netCDF Operators version 4.9.5 (Homepage = http://nco.sf.net, Code = http://github.com/nco/nco)
- NUM_LAND_CAT :
- 21
- OBS_NUDGE_OPT :
- 0
- OPT_ALB :
- 2
- OPT_BTR :
- 2
- OPT_CRS :
- 1
- OPT_FRZ :
- 1
- OPT_GLA :
- 1
- OPT_INF :
- 1
- OPT_RAD :
- 3
- OPT_RSF :
- 1
- OPT_RUN :
- 5
- OPT_SFC :
- 1
- OPT_SNF :
- 4
- OPT_STC :
- 3
- OPT_TBOT :
- 1
- PARENT_GRID_RATIO :
- 1
- PARENT_ID :
- 0
- POLE_LAT :
- 90.0
- POLE_LON :
- 0.0
- PREC_ACC_DT :
- 60.0
- Project :
- USGS CONUS404
- RADT :
- 5.0
- RA_LW_PHYSICS :
- 4
- RA_SW_PHYSICS :
- 4
- SCALAR_ADV_OPT :
- 1
- SCALAR_PBLMIX :
- 0
- SF_LAKE_PHYSICS :
- 0
- SF_OCEAN_PHYSICS :
- 0
- SF_SFCLAY_PHYSICS :
- 1
- SF_SURFACE_PHYSICS :
- 4
- SF_URBAN_PHYSICS :
- 0
- SGFDDA_END_H :
- 0
- SGFDDA_INTERVAL_M :
- 0
- SHCU_PHYSICS :
- 0
- SIMULATION_INITIALIZATION_TYPE :
- REAL-DATA CASE
- SIMULATION_START_DATE :
- 1979-10-01_00:00:00
- SKEBS_ON :
- 0
- SMOOTH_OPTION :
- 2
- SOUTH-NORTH_GRID_DIMENSION :
- 1016
- SOUTH-NORTH_PATCH_END_STAG :
- 1016
- SOUTH-NORTH_PATCH_END_UNSTAG :
- 1015
- SOUTH-NORTH_PATCH_START_STAG :
- 1
- SOUTH-NORTH_PATCH_START_UNSTAG :
- 1
- SPEC_BDY_FINAL_MU :
- 1
- SST_UPDATE :
- 1
- STAND_LON :
- -97.9000015258789
- START_DATE :
- 1979-10-01_00:00:00
- SURFACE_INPUT_SOURCE :
- 1
- SWINT_OPT :
- 0
- SWRAD_SCAT :
- 1.0
- Source_Code :
- make_conusii_2d.csh
- TITLE :
- OUTPUT FROM WRF V3.9.1.1 MODEL
- TKE_ADV_OPT :
- 1
- TOPO_WIND :
- 1
- TRACER_PBLMIX :
- 1
- TRUELAT1 :
- 30.0
- TRUELAT2 :
- 50.0
- USE_Q_DIABATIC :
- 0
- USE_THETA_M :
- 0
- WEST-EAST_GRID_DIMENSION :
- 1368
- WEST-EAST_PATCH_END_STAG :
- 1368
- WEST-EAST_PATCH_END_UNSTAG :
- 1367
- WEST-EAST_PATCH_START_STAG :
- 1
- WEST-EAST_PATCH_START_UNSTAG :
- 1
- W_DAMPING :
- 1
- YSU_TOPDOWN_PBLMIX :
- 0
- history :
- Tue Mar 29 16:35:22 2022: ncrcat -A -vW /glade/scratch/kyoko/USGS/conus404_production_outputs/GHT/WY1980/CONUS404_W_d01_1979-10-01_00:00:00.nc /glade/scratch/kyoko/USGS/conus404_production_outputs/OUTPUT/WY1980/wrf2d_d01_1979-10-01_00:00:00 Tue Mar 29 16:35:21 2022: ncrcat -A -vZ /glade/scratch/kyoko/USGS/conus404_production_outputs/GHT/WY1980/CONUS404_Z_d01_1979-10-01_00:00:00.nc /glade/scratch/kyoko/USGS/conus404_production_outputs/OUTPUT/WY1980/wrf2d_d01_1979-10-01_00:00:00
- history_of_appended_files :
- Tue Mar 29 16:35:22 2022: Appended file /glade/scratch/kyoko/USGS/conus404_production_outputs/GHT/WY1980/CONUS404_W_d01_1979-10-01_00:00:00.nc had no "history" attribute Tue Mar 29 16:35:21 2022: Appended file /glade/scratch/kyoko/USGS/conus404_production_outputs/GHT/WY1980/CONUS404_Z_d01_1979-10-01_00:00:00.nc had no "history" attribute
Conus404 projection
c404_proj = "+proj=lcc +lat_0=39.1000061035156 +lon_0=-97.9000015258789 +lat_1=30 +lat_2=50 +x_0=0 +y_0=0 +R=6370000 +units=m +no_defs"GDPTools
In the following cells GDPtools if first used to generate a weighting file used for area-weighted statistics and secondly to apply the weighting file to interpolate the Conus404 gridded data to each of the HUC12 polygons using an area-weighted mean. GDPTools has a number of data classes to parameterize the input data, which can be though of as the source and target data with some accompanying attributes of each. In this case we use the UserCatData class.
Source Data: - ds: Xarray dataset - x_coord: Name of x-coordinate - y_cord: Name of y-coordinate - t_coord: Name of t-coordinate - proj_ds: The source data’s projection. This can be anything that can by read by pyproj.crs.CRS.from_user_input() method. - var: List of variables in the xarray dataset that will be processed.
Target Data: - f_feature: The GeoDataFrame - In this case working_gdf define above. - proj_feature: Similar to proj_ds above but for the Target GeoDataFrame. - id_feature: The column heading of the feature used to identify the target for the interpolation - In this case “huc12”
Period: The period of interest. This is where things can go sideways. It’s always a good strategy to start with a small slice of data and work up. For large Target datasets, for example HUC12s for all of CONUS, you would quickly run out of memory if processing more that a year at a time. Also some servers have a limit on the size of the data requested. In this case we have a small example - so it’s no issue.
data_crs = c404_proj
x_coord = "x"
y_coord = "y"
t_coord = "time"
sdate = "1979-10-01T00:00:00.000000000"
edate = "1979-10-07T00:00:00.000000000"
var = ["T2", "ACRAINLSM"]
shp_crs = 4326
shp_poly_idx = "huc12"
wght_gen_crs = 6931
user_data = UserCatData(
ds=ds,
proj_ds=data_crs,
x_coord=x_coord,
y_coord=y_coord,
t_coord=t_coord,
var=var,
f_feature=working_gdf,
proj_feature=shp_crs,
id_feature=shp_poly_idx,
period=[sdate, edate],
)Generate the weights
Generate the weights and save to a file for later use.
genwghts = True
wght_gen = WeightGen(
user_data=user_data,
method="serial",
output_file="wghts_drb_ser_c404daily.csv",
weight_gen_crs=6931
)
if genwghts:
wdf = wght_gen.calculate_weights()Using serial engine
Generating grid-cell polygons finished in 0.3 second(s)
Data preparation finished in 0.3227 seconds
Reprojecting to epsg:EPSG:6931 finished in 0.05 second(s)
Validating polygons
- validating source polygons
- fixing 0 invalid polygons.
- validating target polygons
- fixing 0 invalid polygons.
Validate polygons finished in 0.0270 seconds
Intersections finished in 0.4099 seconds
Weight gen finished in 0.4377 secondsAggregate the data using the weights generated above.
agg_gen = AggGen(
user_data=user_data,
stat_method="masked_mean",
agg_engine="serial",
agg_writer="netcdf",
weights='wghts_drb_ser_c404daily.csv',
out_path='.',
file_prefix="testing_p"
)
ngdf, ds_out = agg_gen.calculate_agg()Processing: T2
Data prepped for aggregation in 0.0028 seconds
Data aggregated in 2.7784 seconds
Processing: ACRAINLSM
Data prepped for aggregation in 0.0014 seconds
Data aggregated in 1.0863 seconds
Saving netcdf file to testing_p.ncGenerate a plot of the resulting interpolatoin
aggdata = agg_gen.agg_data.get('T2').da
aggdata<xarray.DataArray 'T2' (time: 7, y: 105, x: 58)> Size: 171kB
dask.array<getitem, shape=(7, 105, 58), dtype=float32, chunksize=(7, 105, 58), chunktype=numpy.ndarray>
Coordinates:
lat (y, x) float32 24kB dask.array<chunksize=(105, 58), meta=np.ndarray>
lon (y, x) float32 24kB dask.array<chunksize=(105, 58), meta=np.ndarray>
* time (time) datetime64[ns] 56B 1979-10-01 1979-10-02 ... 1979-10-07
* x (x) float64 464B 1.744e+06 1.748e+06 ... 1.968e+06 1.972e+06
* y (y) float64 840B 2.08e+05 2.12e+05 2.16e+05 ... 6.2e+05 6.24e+05
metpy_crs object 8B Projection: lambert_conformal_conic
Attributes:
description: TEMP at 2 M
grid_mapping: crs
long_name: Temperature at 2 meters
units: Karray(['1979-10-01T00:00:00.000000000', '1979-10-02T00:00:00.000000000',
'1979-10-03T00:00:00.000000000', '1979-10-04T00:00:00.000000000',
'1979-10-05T00:00:00.000000000', '1979-10-06T00:00:00.000000000',
'1979-10-07T00:00:00.000000000'], dtype='datetime64[ns]')- long_name :
- x coordinate of projection
- standard_name :
- projection_x_coordinate
- units :
- meters
array([1744000., 1748000., 1752000., 1756000., 1760000., 1764000., 1768000.,
1772000., 1776000., 1780000., 1784000., 1788000., 1792000., 1796000.,
1800000., 1804000., 1808000., 1812000., 1816000., 1820000., 1824000.,
1828000., 1832000., 1836000., 1840000., 1844000., 1848000., 1852000.,
1856000., 1860000., 1864000., 1868000., 1872000., 1876000., 1880000.,
1884000., 1888000., 1892000., 1896000., 1900000., 1904000., 1908000.,
1912000., 1916000., 1920000., 1924000., 1928000., 1932000., 1936000.,
1940000., 1944000., 1948000., 1952000., 1956000., 1960000., 1964000.,
1968000., 1972000.])- long_name :
- y coordinate of projection
- standard_name :
- projection_y_coordinate
- units :
- meters
array([208000., 212000., 216000., 220000., 224000., 228000., 232000., 236000.,
240000., 244000., 248000., 252000., 256000., 260000., 264000., 268000.,
272000., 276000., 280000., 284000., 288000., 292000., 296000., 300000.,
304000., 308000., 312000., 316000., 320000., 324000., 328000., 332000.,
336000., 340000., 344000., 348000., 352000., 356000., 360000., 364000.,
368000., 372000., 376000., 380000., 384000., 388000., 392000., 396000.,
400000., 404000., 408000., 412000., 416000., 420000., 424000., 428000.,
432000., 436000., 440000., 444000., 448000., 452000., 456000., 460000.,
464000., 468000., 472000., 476000., 480000., 484000., 488000., 492000.,
496000., 500000., 504000., 508000., 512000., 516000., 520000., 524000.,
528000., 532000., 536000., 540000., 544000., 548000., 552000., 556000.,
560000., 564000., 568000., 572000., 576000., 580000., 584000., 588000.,
592000., 596000., 600000., 604000., 608000., 612000., 616000., 620000.,
624000.])array(<metpy.plots.mapping.CFProjection object at 0x16472cd90>,
dtype=object)- timePandasIndex
PandasIndex(DatetimeIndex(['1979-10-01', '1979-10-02', '1979-10-03', '1979-10-04', '1979-10-05', '1979-10-06', '1979-10-07'], dtype='datetime64[ns]', name='time', freq=None)) - xPandasIndex
PandasIndex(Index([1744000.0, 1748000.0, 1752000.0, 1756000.0, 1760000.0, 1764000.0, 1768000.0, 1772000.0, 1776000.0, 1780000.0, 1784000.0, 1788000.0, 1792000.0, 1796000.0, 1800000.0, 1804000.0, 1808000.0, 1812000.0, 1816000.0, 1820000.0, 1824000.0, 1828000.0, 1832000.0, 1836000.0, 1840000.0, 1844000.0, 1848000.0, 1852000.0, 1856000.0, 1860000.0, 1864000.0, 1868000.0, 1872000.0, 1876000.0, 1880000.0, 1884000.0, 1888000.0, 1892000.0, 1896000.0, 1900000.0, 1904000.0, 1908000.0, 1912000.0, 1916000.0, 1920000.0, 1924000.0, 1928000.0, 1932000.0, 1936000.0, 1940000.0, 1944000.0, 1948000.0, 1952000.0, 1956000.0, 1960000.0, 1964000.0, 1968000.0, 1972000.0], dtype='float64', name='x')) - yPandasIndex
PandasIndex(Index([208000.0000000042, 212000.0000000042, 216000.0000000042, 220000.0000000042, 224000.0000000042, 228000.0000000042, 232000.0000000042, 236000.0000000042, 240000.0000000042, 244000.0000000042, ... 588000.0000000042, 592000.0000000042, 596000.0000000042, 600000.0000000042, 604000.0000000042, 608000.0000000042, 612000.0000000042, 616000.0000000042, 620000.0000000042, 624000.0000000042], dtype='float64', name='y', length=105))
- description :
- TEMP at 2 M
- grid_mapping :
- crs
- long_name :
- Temperature at 2 meters
- units :
- K
wvar = 'T2'
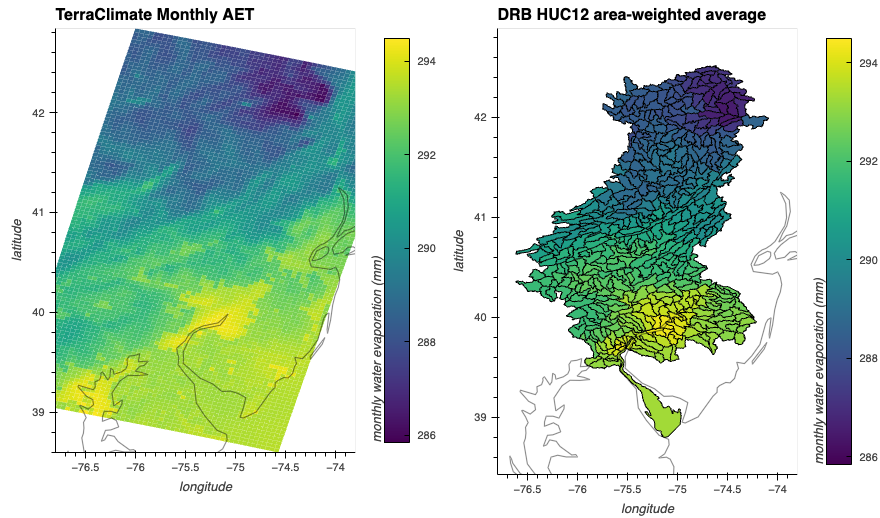
time_step = "1979-10-03T00:00:00.000000000"vmin = np.nanmin(aggdata.sel(time=time_step).values)
vmax = np.nanmax(aggdata.sel(time=time_step).values)
print(vmin, vmax)285.8411 294.49335ngdf['T2'] = ds_out[wvar].sel(time=time_step)
c404_agg = ngdf.to_crs(4326).hvplot(
c='T2', geo=True, coastline='50m', frame_width=300, alpha=1, cmap='viridis', clim=(vmin, vmax),
xlabel="longitude", ylabel="latitude", clabel="monthly water evaporation (mm)", xlim=(-76.8, -73.8),
title="DRB HUC12 area-weighted average", aspect='equal'
)
c404_raw = aggdata.sel(time=time_step).hvplot.quadmesh(
'lon', 'lat', 'T2', cmap='viridis', alpha=1, grid=True, geo=True, coastline='50m', frame_width=300, clim=(vmin,vmax),
xlabel="longitude", ylabel="latitude", clabel="monthly water evaporation (mm)", xlim=(-76.8, -73.8),
title="TerraClimate Monthly AET", aspect='equal'
)#| vscode: {languageId: python}
c404_raw + c404_agg